Difference between revisions of "PpsE"
(→This gene is a member of the following regulons) |
|||
| Line 54: | Line 54: | ||
=== Additional information=== | === Additional information=== | ||
| − | + | * A mutation was found in this gene after evolution under relaxed selection for sporulation {{PubMed|21821766}} | |
| − | |||
| Line 80: | Line 79: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' | + | * '''[[SubtInteract|Interactions]]:''' |
| − | * '''Localization:''' cell membrane (according to Swiss-Prot) | + | * '''[[Localization]]:''' cell membrane (according to Swiss-Prot) |
=== Database entries === | === Database entries === | ||
| Line 128: | Line 127: | ||
=References= | =References= | ||
'''Additional publications:''' {{PubMed|20817675}} | '''Additional publications:''' {{PubMed|20817675}} | ||
| − | <pubmed>10471562, 9387222, </pubmed> | + | <pubmed>10471562, 9387222, 21821766</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 12:02, 15 August 2011
- Description: plipastatin synthetase
| Gene name | ppsE |
| Synonyms | |
| Essential | no |
| Product | plipastatin synthetase |
| Function | production of the antibacterial compound plipastatin |
| MW, pI | 144 kDa, 5.659 |
| Gene length, protein length | 3837 bp, 1279 aa |
| Immediate neighbours | yngL, ppsD |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 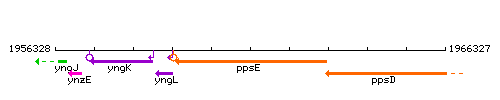
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
miscellaneous metabolic pathways, biosynthesis of antibacterial compounds, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU18300
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
- A mutation was found in this gene after evolution under relaxed selection for sporulation PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: UPF0713 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: O31827
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Christopher T Brown, Laura K Fishwick, Binna M Chokshi, Marissa A Cuff, Jay M Jackson, Travis Oglesby, Alison T Rioux, Enrique Rodriguez, Gregory S Stupp, Austin H Trupp, James S Woollcombe-Clarke, Tracy N Wright, William J Zaragoza, Jennifer C Drew, Eric W Triplett, Wayne L Nicholson
Whole-genome sequencing and phenotypic analysis of Bacillus subtilis mutants following evolution under conditions of relaxed selection for sporulation.
Appl Environ Microbiol: 2011, 77(19);6867-77
[PubMed:21821766]
[WorldCat.org]
[DOI]
(I p)
K Tsuge, T Ano, M Hirai, Y Nakamura, M Shoda
The genes degQ, pps, and lpa-8 (sfp) are responsible for conversion of Bacillus subtilis 168 to plipastatin production.
Antimicrob Agents Chemother: 1999, 43(9);2183-92
[PubMed:10471562]
[WorldCat.org]
[DOI]
(P p)
Valentina Tosato, Alessandra M Albertini, Michela Zotti, Sabrina Sonda, Carlo V Bruschi
Sequence completion, identification and definition of the fengycin operon in Bacillus subtilis 168.
Microbiology (Reading): 1997, 143 ( Pt 11);3443-3450
[PubMed:9387222]
[WorldCat.org]
[DOI]
(P p)