Difference between revisions of "DynA"
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || fusion of membranes | |style="background:#ABCDEF;" align="center"|'''Function''' || fusion of membranes | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http:// | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU22030 dynA] |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/DynA DynA] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/DynA DynA] | ||
| Line 24: | Line 24: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ypbS]]'', ''[[fbpC]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ypbS]]'', ''[[fbpC]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU22030 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU22030 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU22030 Advanced_DNA] |
|- | |- | ||
|- | |- | ||
Revision as of 13:06, 13 May 2013
- Description: dynamin-like protein, mediates membrane fusion
| Gene name | dynA |
| Synonyms | ypbR |
| Essential | no |
| Product | dynamin-like protein |
| Function | fusion of membranes |
| Gene expression levels in SubtiExpress: dynA | |
| Interactions involving this protein in SubtInteract: DynA | |
| MW, pI | 137 kDa, 5.724 |
| Gene length, protein length | 3579 bp, 1193 aa |
| Immediate neighbours | ypbS, fbpC |
| Sequences | Protein DNA Advanced_DNA |
Genetic context 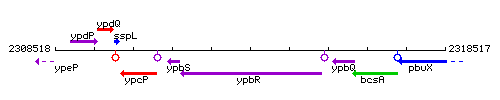
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed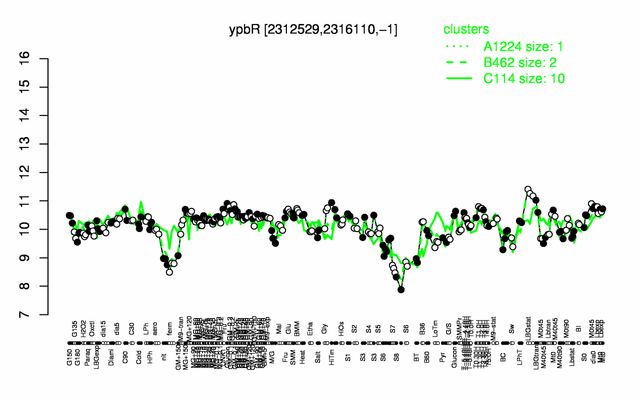
| |
Contents
Categories containing this gene/protein
cell division, cell shape, membrane dynamics, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU22030
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: mediates nucleotide independent membrane fusion in vitro PubMed
- Protein family: gerABKA family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains: two separate dynamin-like subunits and GTPase domains PubMed
- Modification:
- Cofactor(s): Mg(2+) PubMed
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P54159
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: dynA PubMed
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications
Felix Dempwolff, Hanna M Wischhusen, Mara Specht, Peter L Graumann
The deletion of bacterial dynamin and flotillin genes results in pleiotrophic effects on cell division, cell growth and in cell shape maintenance.
BMC Microbiol: 2012, 12;298
[PubMed:23249255]
[WorldCat.org]
[DOI]
(I e)
Frank Bürmann, Prachi Sawant, Marc Bramkamp
Identification of interaction partners of the dynamin-like protein DynA from Bacillus subtilis.
Commun Integr Biol: 2012, 5(4);362-9
[PubMed:23060960]
[WorldCat.org]
[DOI]
(I p)
Frank Bürmann, Nina Ebert, Suey van Baarle, Marc Bramkamp
A bacterial dynamin-like protein mediating nucleotide-independent membrane fusion.
Mol Microbiol: 2011, 79(5);1294-304
[PubMed:21205012]
[WorldCat.org]
[DOI]
(I p)
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
L Chen, L P James, J D Helmann
Metalloregulation in Bacillus subtilis: isolation and characterization of two genes differentially repressed by metal ions.
J Bacteriol: 1993, 175(17);5428-37
[PubMed:8396117]
[WorldCat.org]
[DOI]
(P p)