Difference between revisions of "CzrA"
| (27 intermediate revisions by 6 users not shown) | |||
| Line 1: | Line 1: | ||
| − | * '''Description:''' transcriptional repressor of cadA and czcD <br/><br/> | + | * '''Description:''' transcriptional repressor ([[transcription factors of the ArsR family|ArsR family]]), ''[[cadA]]'' and ''[[czcD]]'' <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || transcriptional repressor (ArsR family) | + | |style="background:#ABCDEF;" align="center"| '''Product''' || transcriptional repressor ([[transcription factors of the ArsR family|ArsR family]]) |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || regulation of resistance against toxic metal cations | |style="background:#ABCDEF;" align="center"|'''Function''' || regulation of resistance against toxic metal cations | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU19120 czrA] | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://cellpublisher.gobics.de/view/GqbblRJkBH metal ion homeostasis]''' | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 12 kDa, 5.719 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 12 kDa, 5.719 | ||
| Line 20: | Line 24: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yobW]]'', ''[[yocA]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yobW]]'', ''[[yocA]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU19120 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU19120 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU19120 DNA_with_flanks] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:yozA_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:yozA_context.gif]] | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
| + | |- | ||
| + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=czrA_2084786_2085109_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:czrA_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU19120]] | ||
|- | |- | ||
|} | |} | ||
__TOC__ | __TOC__ | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/> | ||
| − | + | = [[Categories]] containing this gene/protein = | |
| + | {{SubtiWiki category|[[trace metal homeostasis (Cu, Zn, Ni, Mn, Mo)]]}}, | ||
| + | {{SubtiWiki category|[[transcription factors and their control]]}}, | ||
| + | {{SubtiWiki category|[[resistance against toxic metals]]}} | ||
| + | |||
| + | = This gene is a member of the following [[regulons]] = | ||
| + | =The [[CzrA regulon]]:= | ||
| + | * ''[[cadA]]'', ''[[czcD]]-[[trkA]]'' | ||
=The gene= | =The gene= | ||
| Line 35: | Line 52: | ||
=== Basic information === | === Basic information === | ||
| − | * ''' | + | * '''Locus tag:''' BSU19120 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU19120&redirect=T BSU19120] | ||
* '''DBTBS entry:''' no entry | * '''DBTBS entry:''' no entry | ||
| Line 46: | Line 64: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
=The protein= | =The protein= | ||
| Line 54: | Line 71: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
| − | * '''Protein family:''' | + | * '''Protein family:''' [[transcription factors of the ArsR family|ArsR family]] |
* '''Paralogous protein(s):''' | * '''Paralogous protein(s):''' | ||
| Line 68: | Line 85: | ||
* '''Cofactor(s):''' | * '''Cofactor(s):''' | ||
| − | * '''Effectors of protein activity:''' | + | * '''Effectors of protein activity:''' |
| + | ** DNA-binding activity of CzrA is inactivated by high concentrations of toxic metal ions (induction) | ||
| − | * '''Interactions:''' | + | * '''[[SubtInteract|Interactions]]:''' |
| − | * '''Localization:''' | + | * '''[[Localization]]:''' |
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU19120&redirect=T BSU19120] | ||
* '''Structure:''' | * '''Structure:''' | ||
| + | ** [http://www.rcsb.org/pdb/explore/explore.do?structureId=1R1U 1R1U] (the apo-repressor from ''Staph. aureus'', 49% identity, 82% similarity) {{PubMed|14568530}} | ||
| + | ** [http://www.rcsb.org/pdb/explore/explore.do?structureId=1R1V 1R1V] (the Zn(II) form from ''Staph. aureus'', 49% identity, 82% similarity) {{PubMed|14568530}} | ||
| + | ** [http://www.rcsb.org/pdb/explore/explore.do?structureId=2KJB 2KJB] (the DNA-bound form from ''Staph. aureus'', 49% identity, 82% similarity) {{PubMed|19822742}} | ||
| + | * '''UniProt:''' [http://www.uniprot.org/uniprot/O31844 O31844] | ||
| − | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU19120] | |
| − | |||
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu | ||
* '''E.C. number:''' | * '''E.C. number:''' | ||
| Line 90: | Line 111: | ||
* '''Operon:''' | * '''Operon:''' | ||
| − | * '''[ | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=czrA_2084786_2085109_-1 czrA] {{PubMed|22383849}} |
| + | |||
| + | * '''Sigma factor:''' | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 96: | Line 119: | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
| Line 117: | Line 140: | ||
=References= | =References= | ||
| + | <pubmed>15948947, 16430705, 14568530 19822742 25213752 22007899</pubmed> | ||
| − | + | [[Category:Protein-coding genes]] | |
Latest revision as of 09:24, 19 September 2014
- Description: transcriptional repressor (ArsR family), cadA and czcD
| Gene name | czrA |
| Synonyms | yozA |
| Essential | no |
| Product | transcriptional repressor (ArsR family) |
| Function | regulation of resistance against toxic metal cations |
| Gene expression levels in SubtiExpress: czrA | |
| Metabolic function and regulation of this protein in SubtiPathways: metal ion homeostasis | |
| MW, pI | 12 kDa, 5.719 |
| Gene length, protein length | 321 bp, 107 aa |
| Immediate neighbours | yobW, yocA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 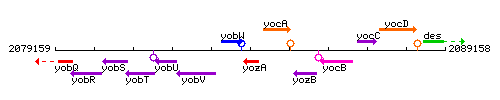
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed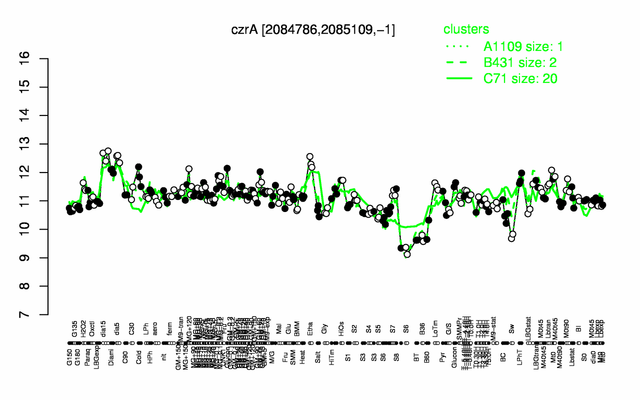
| |
Contents
Categories containing this gene/protein
trace metal homeostasis (Cu, Zn, Ni, Mn, Mo), transcription factors and their control, resistance against toxic metals
This gene is a member of the following regulons
The CzrA regulon:
The gene
Basic information
- Locus tag: BSU19120
Phenotypes of a mutant
Database entries
- BsubCyc: BSU19120
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ArsR family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- DNA-binding activity of CzrA is inactivated by high concentrations of toxic metal ions (induction)
Database entries
- BsubCyc: BSU19120
- Structure:
- UniProt: O31844
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Zhen Ma, Pete Chandrangsu, Tyler C Helmann, Adisak Romsang, Ahmed Gaballa, John D Helmann
Bacillithiol is a major buffer of the labile zinc pool in Bacillus subtilis.
Mol Microbiol: 2014, 94(4);756-70
[PubMed:25213752]
[WorldCat.org]
[DOI]
(I p)
Dhruva K Chakravorty, Bing Wang, Chul Won Lee, David P Giedroc, Kenneth M Merz
Simulations of allosteric motions in the zinc sensor CzrA.
J Am Chem Soc: 2012, 134(7);3367-76
[PubMed:22007899]
[WorldCat.org]
[DOI]
(I p)
Alphonse I Arunkumar, Gregory C Campanello, David P Giedroc
Solution structure of a paradigm ArsR family zinc sensor in the DNA-bound state.
Proc Natl Acad Sci U S A: 2009, 106(43);18177-82
[PubMed:19822742]
[WorldCat.org]
[DOI]
(I p)
Duncan R Harvie, Claudia Andreini, Gabriele Cavallaro, Wenmao Meng, Bernard A Connolly, Ken-ichi Yoshida, Yasutaro Fujita, Colin R Harwood, David S Radford, Stephen Tottey, Jennifer S Cavet, Nigel J Robinson
Predicting metals sensed by ArsR-SmtB repressors: allosteric interference by a non-effector metal.
Mol Microbiol: 2006, 59(4);1341-56
[PubMed:16430705]
[WorldCat.org]
[DOI]
(P p)
Charles M Moore, Ahmed Gaballa, Monica Hui, Rick W Ye, John D Helmann
Genetic and physiological responses of Bacillus subtilis to metal ion stress.
Mol Microbiol: 2005, 57(1);27-40
[PubMed:15948947]
[WorldCat.org]
[DOI]
(P p)
Christoph Eicken, Mario A Pennella, Xiaohua Chen, Karl M Koshlap, Michael L VanZile, James C Sacchettini, David P Giedroc
A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J Mol Biol: 2003, 333(4);683-95
[PubMed:14568530]
[WorldCat.org]
[DOI]
(P p)