Difference between revisions of "CwlO"
| Line 135: | Line 135: | ||
=Biological materials = | =Biological materials = | ||
| − | * '''Mutant:''' | + | * '''Mutant:''' BKE34800 (''[[cwlO]]''::''erm'', available in the BGSC and in [[Jörg Stülke]]'s lab) |
* '''Expression vector:''' | * '''Expression vector:''' | ||
Revision as of 18:59, 1 February 2015
- Description: D,L-endopeptidase-type autolysin, primary autolytic pathway for cell elongation
| Gene name | cwlO |
| Synonyms | yzkA, yvcE |
| Essential | no |
| Product | endopeptidase-type autolysin |
| Function | cell wall synthesis, cell elongation |
| Gene expression levels in SubtiExpress: cwlO | |
| Interactions involving this protein in SubtInteract: CwlO | |
| MW, pI | 50 kDa, 5.326 |
| Gene length, protein length | 1419 bp, 473 aa |
| Immediate neighbours | trxB, yvcD |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 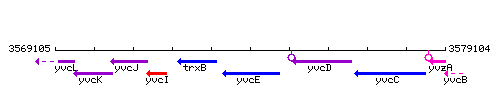
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed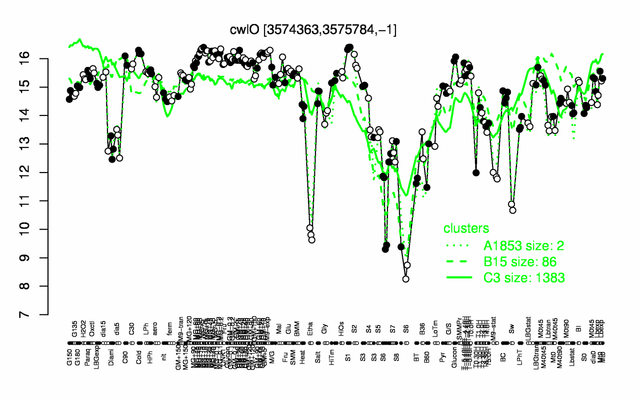
| |
Contents
Categories containing this gene/protein
cell wall synthesis, cell wall degradation/ turnover
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU34800
Phenotypes of a mutant
- a cwlO lytE mutant is not viable PubMed
- shorter, fatter cells, this can be rescued by addition of Mg(2+) PubMed
Database entries
- BsubCyc: BSU34800
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: peptidase C40 family (according to Swiss-Prot)
- Paralogous protein(s): the C-terminal D,L-endopeptidase domains of LytE, LytF, CwlS, and CwlO exhibit strong sequence similarity
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- BsubCyc: BSU34800
- Structure:
- UniProt: P40767
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Additional information:
- The mRNA has a long 5' leader region (187 N). This may indicate RNA-based regulation PubMed
- the leader mRNA is subject to cleavage by RNase Y, this results in limited expression of cwlO PubMed
- there is a SigB-dependent anti-sense transcript which is expressed in the stationary phase PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 207 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 200 PubMed
Biological materials
- Mutant: BKE34800 (cwlO::erm, available in the BGSC and in Jörg Stülke's lab)
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications
David Noone, Letal I Salzberg, Eric Botella, Katrin Bäsell, Dörte Becher, Haike Antelmann, Kevin M Devine
A highly unstable transcript makes CwlO D,L-endopeptidase expression responsive to growth conditions in Bacillus subtilis.
J Bacteriol: 2014, 196(2);237-47
[PubMed:24163346]
[WorldCat.org]
[DOI]
(I p)
Patricia Domínguez-Cuevas, Ida Porcelli, Richard A Daniel, Jeff Errington
Differentiated roles for MreB-actin isologues and autolytic enzymes in Bacillus subtilis morphogenesis.
Mol Microbiol: 2013, 89(6);1084-98
[PubMed:23869552]
[WorldCat.org]
[DOI]
(I p)
Jeffrey Meisner, Paula Montero Llopis, Lok-To Sham, Ethan Garner, Thomas G Bernhardt, David Z Rudner
FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis.
Mol Microbiol: 2013, 89(6);1069-83
[PubMed:23855774]
[WorldCat.org]
[DOI]
(I p)
Masayuki Hashimoto, Seika Ooiwa, Junichi Sekiguchi
Synthetic lethality of the lytE cwlO genotype in Bacillus subtilis is caused by lack of D,L-endopeptidase activity at the lateral cell wall.
J Bacteriol: 2012, 194(4);796-803
[PubMed:22139507]
[WorldCat.org]
[DOI]
(I p)
Nobuo Mitsui, Hisashi Murasawa, Junichi Sekiguchi
Disruption of the cell wall lytic enzyme CwlO affects the amount and molecular size of poly-γ-glutamic acid produced by Bacillus subtilis (natto).
J Gen Appl Microbiol: 2011, 57(1);35-43
[PubMed:21478646]
[WorldCat.org]
[DOI]
(I p)
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
Paola Bisicchia, David Noone, Efthimia Lioliou, Alistair Howell, Sarah Quigley, Thomas Jensen, Hanne Jarmer, Kevin M Devine
The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis.
Mol Microbiol: 2007, 65(1);180-200
[PubMed:17581128]
[WorldCat.org]
[DOI]
(P p)
Hiroyuki Yamaguchi, Kazumi Furuhata, Tatsuya Fukushima, Hiroki Yamamoto, Junichi Sekiguchi
Characterization of a new Bacillus subtilis peptidoglycan hydrolase gene, yvcE (named cwlO), and the enzymatic properties of its encoded protein.
J Biosci Bioeng: 2004, 98(3);174-81
[PubMed:16233686]
[WorldCat.org]
[DOI]
(P p)