Difference between revisions of "YvyD"
(→Expression and regulation) |
|||
| Line 44: | Line 44: | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| + | {{SubtiWiki regulon|[[PhoP regulon]]}}, | ||
{{SubtiWiki regulon|[[SigB regulon]]}}, | {{SubtiWiki regulon|[[SigB regulon]]}}, | ||
{{SubtiWiki regulon|[[SigH regulon]]}} | {{SubtiWiki regulon|[[SigH regulon]]}} | ||
| Line 147: | Line 148: | ||
=References= | =References= | ||
| − | <pubmed>11948165,9852014,10913081,22517742,12107147 15805528 22950019 22582280</pubmed> | + | <pubmed>11948165,9852014,10913081,22517742,12107147 15805528 22950019 22582280 25666134</pubmed> |
==Publications on the homologous proteins from other bacteria== | ==Publications on the homologous proteins from other bacteria== | ||
<pubmed> 22605777 24279750 </pubmed> | <pubmed> 22605777 24279750 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 13:06, 11 February 2015
- Description: general stress protein, required for ribosome dimerization in the stationary phase, required for protection against paraquat stress
| Gene name | yvyD |
| Synonyms | yviI |
| Essential | no |
| Product | unknown |
| Function | dimerization of ribosomes in the stationary phase, protection against paraquat stress |
| Gene expression levels in SubtiExpress: yvyD | |
| MW, pI | 21 kDa, 5.184 |
| Gene length, protein length | 567 bp, 189 aa |
| Immediate neighbours | secA, smiA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 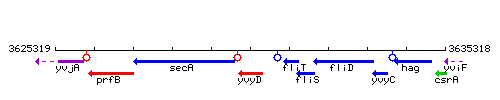
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
general stress proteins (controlled by SigB), resistance against oxidative and electrophile stress, membrane proteins, phosphoproteins
This gene is a member of the following regulons
PhoP regulon, SigB regulon, SigH regulon
The gene
Basic information
- Locus tag: BSU35310
Phenotypes of a mutant
- the mutant is cold-sensitive
- delayed recovery from stationary phase and delayed germination
Database entries
- BsubCyc: BSU35310
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ribosomal protein S30Ae family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- phosphorylated on Arg-6 PubMed
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
- membrane protein
Database entries
- BsubCyc: BSU35310
- Structure:
- UniProt: P28368
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: yvyD PubMed
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Publications on the homologous proteins from other bacteria