Difference between revisions of "SigD"
(→Expression and regulation) |
|||
| Line 60: | Line 60: | ||
* Inactivation of ''[[alsR]]'', ''[[sigD]]'' and ''[[sigW]]'' increases competitive fitness of ''Bacillus subtilis'' under non-sporulating conditions {{PubMed|22344650}} | * Inactivation of ''[[alsR]]'', ''[[sigD]]'' and ''[[sigW]]'' increases competitive fitness of ''Bacillus subtilis'' under non-sporulating conditions {{PubMed|22344650}} | ||
* mucoid phenotype due to the overproduction of poly-gamma-glutamate (because of the loss of ''[[motA]]-[[motB]]'' expression) {{PubMed|24296669}} | * mucoid phenotype due to the overproduction of poly-gamma-glutamate (because of the loss of ''[[motA]]-[[motB]]'' expression) {{PubMed|24296669}} | ||
| + | * not essential for pellicle biofilm formation, but mutant is outcompeted by the wild-type strain when competed during pellicle formation {{PubMed|26122431}} | ||
=== Database entries === | === Database entries === | ||
| Line 133: | Line 134: | ||
* '''Mutant:''' | * '''Mutant:''' | ||
** 1A716 ( ''sigD''::''cat''), {{PubMed|2832368}}, available at [http://pasture.asc.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1A716&Search=1A716 BGSC] | ** 1A716 ( ''sigD''::''cat''), {{PubMed|2832368}}, available at [http://pasture.asc.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1A716&Search=1A716 BGSC] | ||
| + | ** DS6420 (marker-less in NCIB3610) {{PubMed|22329926}} | ||
| + | ** TB188 ''amyE''::Phy-''sfgfp'' (marker-less in NCIB3610 with constitutive expressed ''sfgfp'') {{PubMed|26122431}} | ||
| + | ** TB204 ''amyE''::Phy-''mKATE2'' (marker-less in NCIB3610 with constitutive expressed ''mKATE2'') {{PubMed|26122431}} | ||
* '''Expression vector:''' | * '''Expression vector:''' | ||
| Line 158: | Line 162: | ||
==Original publications== | ==Original publications== | ||
| − | <pubmed> 22344650 22329926,21097624,22956758 21856853 8955328,7708689,9648743, 8169223,9096206,15175317, 2498284,16357223, 3127378, 9144176,18820022,7602586,2832368,8195064,11987133,,14651647, 9335309 9657996,8157612,15175317 10809682 8932324 20233303 17850253 10207036 16428420 24296669 25099370 24386445</pubmed> | + | <pubmed> 26122431,22344650 22329926,21097624,22956758 21856853 8955328,7708689,9648743, 8169223,9096206,15175317, 2498284,16357223, 3127378, 9144176,18820022,7602586,2832368,8195064,11987133,,14651647, 9335309 9657996,8157612,15175317 10809682 8932324 20233303 17850253 10207036 16428420 24296669 25099370 24386445</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 13:16, 2 July 2015
- Description: RNA polymerase sigma factor SigD
| Gene name | sigD |
| Synonyms | flaB |
| Essential | no |
| Product | RNA polymerase sigma factor SigD |
| Function | regulation of flagella, motility, chemotaxis and autolysis |
| Gene expression levels in SubtiExpress: sigD | |
| Interactions involving this protein in SubtInteract: SigD | |
| MW, pI | 29 kDa, 5.167 |
| Gene length, protein length | 762 bp, 254 aa |
| Immediate neighbours | cheD, swrB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 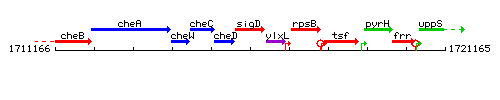
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed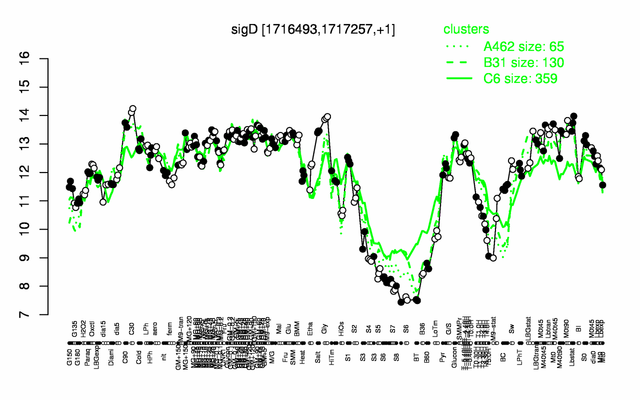
| |
Contents
Categories containing this gene/protein
transcription, sigma factors and their control, motility and chemotaxis
This gene is a member of the following regulons
CodY regulon, DegU regulon, SigD regulon, Spo0A regulon
The SigD regulon
The gene
Basic information
- Locus tag: BSU16470
Phenotypes of a mutant
- Inactivation of alsR, sigD and sigW increases competitive fitness of Bacillus subtilis under non-sporulating conditions PubMed
- mucoid phenotype due to the overproduction of poly-gamma-glutamate (because of the loss of motA-motB expression) PubMed
- not essential for pellicle biofilm formation, but mutant is outcompeted by the wild-type strain when competed during pellicle formation PubMed
Database entries
- BsubCyc: BSU16470
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: sigma-70 factor family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
Database entries
- BsubCyc: BSU16470
- Structure:
- UniProt: P10726
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: part of the fla-che operon
- flgB-flgC-fliE-fliF-fliG-fliH-fliI-fliJ-ylxF-fliK-flgD-flgE-fliL-fliM-fliY-cheY-fliZ-fliP-fliQ-fliR-flhB-flhA-flhF-flhG-cheB-cheA-cheW-cheC-cheD-sigD-swrB PubMed
- ylxF-fliK-flgD-flgE-fliL-fliM-fliY-cheY-fliZ-fliP-fliQ-fliR-flhB-flhA-flhF-flhG-cheB-cheA-cheW-cheC-cheD-sigD-swrB PubMed
- sigD-swrB PubMed
- Regulation: see fla-che operon
- Regulatory mechanism: see fla-che operon
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
The SigD regulon
Original publications