Difference between revisions of "CcpA"
(→References) |
|||
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || mediates carbon catabolite repression (CCR) | |style="background:#ABCDEF;" align="center"|'''Function''' || mediates carbon catabolite repression (CCR) | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbon_flow.html Central C-metabolism]''' | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 36,8 kDa, 5.06 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 36,8 kDa, 5.06 | ||
Revision as of 12:15, 11 June 2009
- Description: Carbon catabolite control protein A, involved in glucose regulation of many genes; represses catabolic genes and activates genes involved in excretion of excess carbon
| Gene name | ccpA |
| Synonyms | graR, alsA, amyR |
| Essential | no |
| Product | transcriptional regulator |
| Function | mediates carbon catabolite repression (CCR) |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 36,8 kDa, 5.06 |
| Gene length, protein length | 1002 bp, 334 amino acids |
| Immediate neighbours | aroA, motP |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 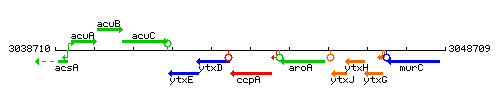
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU29740
Phenotypes of a mutant
Loss of carbon catabolite repression. Loss of PTS-dependent sugar transport due to excessive phosphorylation of HPr by HprK. The mutant is unable to grow on a minimal medium with glucose and ammonium as the only sources of carbon and nitrogen, respectively.
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: transcriptional regulator of carbon catabolite repression (CCR)
- Protein family: LacI family
- Paralogous protein(s):
Genes controlled by CcpA
- Repression by CcpA: abbA, amyE, bglP-bglH, bglS, cccA, citZ-icd-mdh, levD-levE-levF-levG-sacC, licB-licC-licA-licH, phoP-phoR, xylA-xylB, xynP-xynB
Extended information on the protein
- Kinetic information:
- Domains:
- HTH lacI-type Domain (1 – 58)
- DNA binding Domain (6 – 25)
- Modification:
- Cofactor(s): HPr-Ser46-P, Crh-Ser-46-P
- Effectors of protein activity:glucose-6-phosphate, fructose-1,6-bisphosphate Pubmed
- Localization:
Database entries
- Swiss prot entry: P25144
- KEGG entry: [3]
Additional information
Expression and regulation
- Sigma factor:
- Regulation: constitutively expressed PubMed
- Additional information: there are about 3.000 molecules of CcpA per cell PubMed
Biological materials
- Mutant: QB5407 (spc), GP302 (erm), GP300 (an in frame deletion of ccpA), available in Stülke lab
- Expression vector: pGP643 (N-terminal Strep-tag, purification from B. subtilis, for SPINE, in pGP380), available in Stülke lab
- lacZ fusion:
- GFP fusion:
Labs working on this gene/protein
Wolfgang Hillen, Erlangen University, Germany Homepage
Richard Brennan, Houston, Texas, USA Homepage
Milton H. Saier, University of California at San Diego, USA Homepage
Yasutaro Fujita, University of Fukuyama, Japan
Jörg Stülke, University of Göttingen, Germany Homepage
Oscar Kuipers, University of Groningen, The Netherlands Homepage
Your additional remarks
References
Reviews
General and physiological studies
Global analyses (proteome, transcriptome)
Repression of target genes by CcpA
Positive regulation of gene expression by CcpA
Robert P Shivers, Abraham L Sonenshein
Bacillus subtilis ilvB operon: an intersection of global regulons.
Mol Microbiol: 2005, 56(6);1549-59
[PubMed:15916605]
[WorldCat.org]
[DOI]
(P p)
Holger Ludwig, Christoph Meinken, Anastasija Matin, Jörg Stülke
Insufficient expression of the ilv-leu operon encoding enzymes of branched-chain amino acid biosynthesis limits growth of a Bacillus subtilis ccpA mutant.
J Bacteriol: 2002, 184(18);5174-8
[PubMed:12193635]
[WorldCat.org]
[DOI]
(P p)
A J Turinsky, T R Moir-Blais, F J Grundy, T M Henkin
Bacillus subtilis ccpA gene mutants specifically defective in activation of acetoin biosynthesis.
J Bacteriol: 2000, 182(19);5611-4
[PubMed:10986270]
[WorldCat.org]
[DOI]
(P p)
E Presecan-Siedel, A Galinier, R Longin, J Deutscher, A Danchin, P Glaser, I Martin-Verstraete
Catabolite regulation of the pta gene as part of carbon flow pathways in Bacillus subtilis.
J Bacteriol: 1999, 181(22);6889-97
[PubMed:10559153]
[WorldCat.org]
[DOI]
(P p)
A J Turinsky, F J Grundy, J H Kim, G H Chambliss, T M Henkin
Transcriptional activation of the Bacillus subtilis ackA gene requires sequences upstream of the promoter.
J Bacteriol: 1998, 180(22);5961-7
[PubMed:9811655]
[WorldCat.org]
[DOI]
(P p)
F J Grundy, D A Waters, S H Allen, T M Henkin
Regulation of the Bacillus subtilis acetate kinase gene by CcpA.
J Bacteriol: 1993, 175(22);7348-55
[PubMed:8226682]
[WorldCat.org]
[DOI]
(P p)
Control of CcpA activity
CcpA-DNA interaction
Functional analysis of CcpA
Structural analyses
Maria A Schumacher, Gerald Seidel, Wolfgang Hillen, Richard G Brennan
Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors glucose 6-phosphate and fructose 1,6-bisphosphate.
J Mol Biol: 2007, 368(4);1042-50
[PubMed:17376479]
[WorldCat.org]
[DOI]
(P p)
Maria A Schumacher, Gerald Seidel, Wolfgang Hillen, Richard G Brennan
Phosphoprotein Crh-Ser46-P displays altered binding to CcpA to effect carbon catabolite regulation.
J Biol Chem: 2006, 281(10);6793-800
[PubMed:16316990]
[WorldCat.org]
[DOI]
(P p)
Maria A Schumacher, Gregory S Allen, Marco Diel, Gerald Seidel, Wolfgang Hillen, Richard G Brennan
Structural basis for allosteric control of the transcription regulator CcpA by the phosphoprotein HPr-Ser46-P.
Cell: 2004, 118(6);731-41
[PubMed:15369672]
[WorldCat.org]
[DOI]
(P p)