Difference between revisions of "SubtiPathways"
(→Publications describing SubtiWiki and SubtiPathways) |
|||
| (66 intermediate revisions by 7 users not shown) | |||
| Line 1: | Line 1: | ||
| − | [[File:Pathway_start_page_small.png|right|link=http:// | + | [[File:Pathway_start_page_small.png|right|link=http://subtiwiki.uni-goettingen.de/subtipathways/start.php]] |
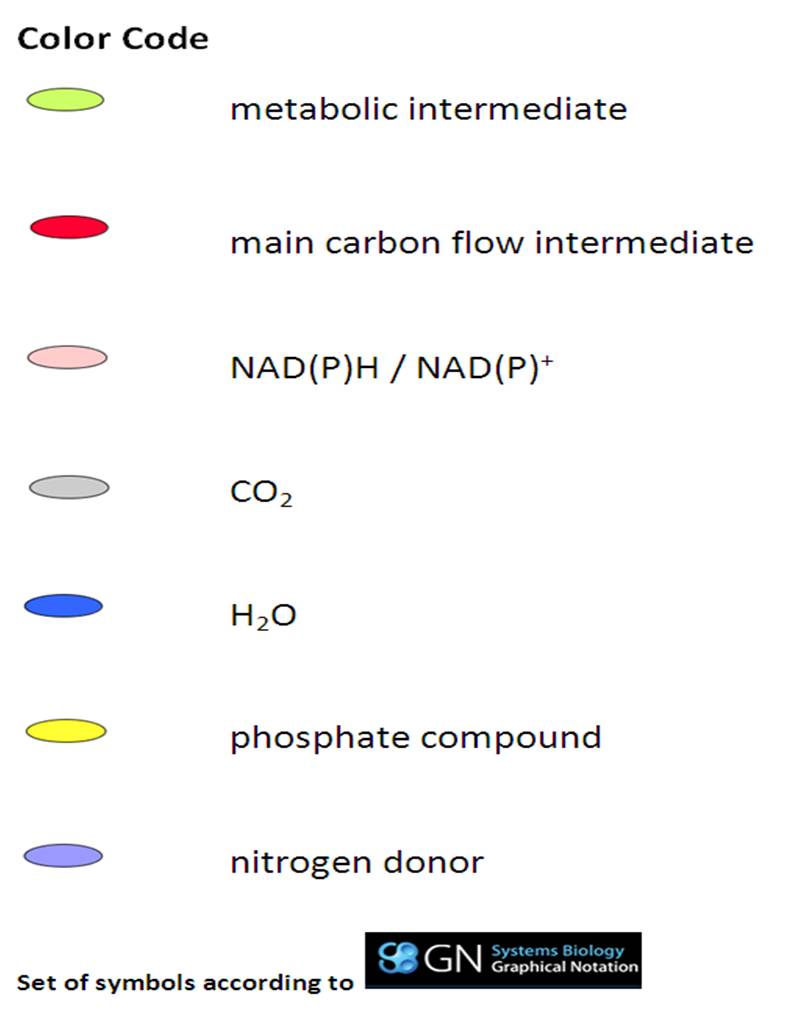
[[File:Legende.jpg|250px|thumb|right|'''legend for ''Subti''Pathways <br/>graphical notation: click to enlarge''']] | [[File:Legende.jpg|250px|thumb|right|'''legend for ''Subti''Pathways <br/>graphical notation: click to enlarge''']] | ||
<big>'''''Subti''Pathways''' is a model of '''''B. subtilis''''' metabolism and regulation in '''SBML/SBGN''' (Systems Biology Markup Language/ Graphical Notation). There is a color code for key metabolites (see the legend on the right side).<br/><br/>It can be used like Google Maps (scroll and zoom to the pathways). The markers of proteins and compounds listed on the right side in each pathway are containing links to the corresponding ''Subti''Wiki and PubChem pages, respectively. There is a short description for each protein in the bubbles, too.</big> | <big>'''''Subti''Pathways''' is a model of '''''B. subtilis''''' metabolism and regulation in '''SBML/SBGN''' (Systems Biology Markup Language/ Graphical Notation). There is a color code for key metabolites (see the legend on the right side).<br/><br/>It can be used like Google Maps (scroll and zoom to the pathways). The markers of proteins and compounds listed on the right side in each pathway are containing links to the corresponding ''Subti''Wiki and PubChem pages, respectively. There is a short description for each protein in the bubbles, too.</big> | ||
| + | <big>'''Contact:'''</big> [http://wwwuser.gwdg.de/~genmibio/stuelke.html General Microbiology, University of Göttingen], [mailto:[email protected] send mail] | ||
| + | |||
| + | <br/> | ||
| + | <big>'''Now online: A description of ''Subti''Wiki, ''Subti''Pathways, and ''Subt''Interact in the 2012 Database issue of Nucleic Acids Research'''</big> | ||
| + | <pubmed> 22096228 </pubmed> | ||
__NOTOC__ | __NOTOC__ | ||
<br/> | <br/> | ||
| − | == A collection of important papers on ''B. subtilis'' [[metabolism]] can be found | + | == A collection of important papers on ''B. subtilis'' [[metabolism]] can be found [[metabolism|here]] == |
<br/> | <br/> | ||
== Carbon metabolism == | == Carbon metabolism == | ||
| − | === [http://subtiwiki.uni-goettingen.de/ | + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=7FvYw Central carbon metabolism] === |
| − | === [http://subtiwiki.uni-goettingen.de/ | + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=ps3UM Utilization of different carbon sources] === |
<br/> | <br/> | ||
| + | |||
== Nitrogen and amino acid metabolism == | == Nitrogen and amino acid metabolism == | ||
| − | === [http://subtiwiki.uni-goettingen.de/ | + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=Cog2y Ammonium assimilation and glutamate metabolism (incl. glutamine and arginine)] === |
| − | === [http://subtiwiki.uni-goettingen.de/ | + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=mONyM Alanine, glycine, serine] === |
| − | === [http://subtiwiki.uni-goettingen.de/ | + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=81K4b Aspartate, asparagine] === |
| − | === [http://subtiwiki.uni-goettingen.de/ | + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=1Jfrj Cysteine, methionine] === |
| − | === [http://subtiwiki.uni-goettingen.de/ | + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=DNohR Histidine] === |
| − | === [http://subtiwiki.uni-goettingen.de/ | + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=V2MSg Isoleucine, valine, leucine] === |
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=lysi Lysine, threonine] === | ||
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=qSUN0 Phenylalanine, tyrosine, tryptophan] === | ||
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=bM03y Proline] === | ||
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=JKtpr Utilization of other nitrogen sources] === | ||
| + | |||
| + | <br/> | ||
| − | == | + | == Lipid metabolism == |
| − | === [http://subtiwiki.uni-goettingen.de/ | + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=fatt Fatty acid metabolism] === |
| − | === [http://subtiwiki.uni-goettingen.de/ | + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=cnliR Fatty acid and phospholipid biosynthesis] === |
<br/> | <br/> | ||
| + | |||
== Nucleotides == | == Nucleotides == | ||
| − | === [http://subtiwiki.uni-goettingen.de/ | + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=nucl Nucleoside catabolism] === |
| − | === [http://subtiwiki.uni-goettingen.de/ | + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=etxDX Purine biosynthesis] === |
| − | === [http://subtiwiki.uni-goettingen.de/ | + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=bxwTq Purine catabolism] === |
| − | === [http://subtiwiki.uni-goettingen.de/ | + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=puri Purine salvage pathways] === |
| − | === [http://subtiwiki.uni-goettingen.de/ | + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=pyri Pyrimidine metabolism] === |
| − | + | <br/> | |
| − | |||
== Metabolism of cofactors == | == Metabolism of cofactors == | ||
| − | === [http://subtiwiki.uni-goettingen.de/ | + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=cUo5h CoA synthesis] === |
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=faSwm Folate biosynthesis] === | ||
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=2GTKN Menaquinone] === | ||
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=4Ww0E Riboflavin and FAD synthesis] === | ||
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=wTyck Thiamin synthesis] === | ||
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=8iQpH Biotin synthesis] === | ||
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=op4I3 Heme biosynthesis] === | ||
| + | |||
| + | ===[http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=wAgIW Molybdopterin synthesis] === | ||
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=3vSpz NAD synthesis] === | ||
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=Zjt4G Lipoate synthesis] === | ||
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=moarx Pyridoxalphospahte synthesis] === | ||
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=AwkEg S-adenosylmethionine synthesis] === | ||
| − | |||
| − | |||
<br/> | <br/> | ||
| + | |||
== Other pathways == | == Other pathways == | ||
| − | === [http://subtiwiki.uni-goettingen.de/ | + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=LlfNo Peptidoglycan synthesis] [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=RqnXf Peptidoglycan catabolism] [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=aARFz Teichonic acid] === |
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=RDamU ppGpp] [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=Tvl8u cdAMP] [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=E6M7Y cdGMP] === | ||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=stre Stress responses] === | ||
| − | === [http://subtiwiki.uni-goettingen.de/pathways/ | + | === [http://subtiwiki.uni-goettingen.de/pathways/phosphorelay.html Phosphorelay] === |
=== [http://subtiwiki.uni-goettingen.de/pathways/cys_meth_and_sulfate_assimilation.html Sulfate assimilation] === | === [http://subtiwiki.uni-goettingen.de/pathways/cys_meth_and_sulfate_assimilation.html Sulfate assimilation] === | ||
| − | === [http://subtiwiki.uni-goettingen.de/ | + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=trna tRNA charging] === |
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=BPSRv Biofilm formation] === | ||
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=95Yqn Protein secretion] === | ||
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=V1JFH Acquisition of iron ] === | ||
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=LNqKW Metal ion homeostasis] === | ||
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=pM5k3 Trace metal homeostasis] === | ||
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=4FfA6 Amino acids transporter] === | ||
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=KheJp Bactoprenol synthesis] === | ||
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=SO2Za Methylglyoxal pathway] === | ||
| + | |||
| + | === [http://subtiwiki.uni-goettingen.de/subtipathways/index.php?pathway=YDHgj Respiration] === | ||
| + | |||
| + | == Publications describing ''Subti''Wiki and ''Subti''Pathways == | ||
| + | <pubmed>19959575 22096228 24178028</pubmed> | ||
Latest revision as of 08:15, 13 May 2014
SubtiPathways is a model of B. subtilis metabolism and regulation in SBML/SBGN (Systems Biology Markup Language/ Graphical Notation). There is a color code for key metabolites (see the legend on the right side).
It can be used like Google Maps (scroll and zoom to the pathways). The markers of proteins and compounds listed on the right side in each pathway are containing links to the corresponding SubtiWiki and PubChem pages, respectively. There is a short description for each protein in the bubbles, too.
Contact: General Microbiology, University of Göttingen, send mail
Now online: A description of SubtiWiki, SubtiPathways, and SubtInteract in the 2012 Database issue of Nucleic Acids Research
Ulrike Mäder, Arne G Schmeisky, Lope A Flórez, Jörg Stülke
SubtiWiki--a comprehensive community resource for the model organism Bacillus subtilis.
Nucleic Acids Res: 2012, 40(Database issue);D1278-87
[PubMed:22096228]
[WorldCat.org]
[DOI]
(I p)
A collection of important papers on B. subtilis metabolism can be found here
Carbon metabolism
Central carbon metabolism
Utilization of different carbon sources
Nitrogen and amino acid metabolism
Ammonium assimilation and glutamate metabolism (incl. glutamine and arginine)
Alanine, glycine, serine
Aspartate, asparagine
Cysteine, methionine
Histidine
Isoleucine, valine, leucine
Lysine, threonine
Phenylalanine, tyrosine, tryptophan
Proline
Utilization of other nitrogen sources
Lipid metabolism
Fatty acid metabolism
Fatty acid and phospholipid biosynthesis
Nucleotides
Nucleoside catabolism
Purine biosynthesis
Purine catabolism
Purine salvage pathways
Pyrimidine metabolism
Metabolism of cofactors
CoA synthesis
Folate biosynthesis
Menaquinone
Riboflavin and FAD synthesis
Thiamin synthesis
Biotin synthesis
Heme biosynthesis
Molybdopterin synthesis
NAD synthesis
Lipoate synthesis
Pyridoxalphospahte synthesis
S-adenosylmethionine synthesis
Other pathways
Peptidoglycan synthesis Peptidoglycan catabolism Teichonic acid
ppGpp cdAMP cdGMP
Stress responses
Phosphorelay
Sulfate assimilation
tRNA charging
Biofilm formation
Protein secretion
Acquisition of iron
Metal ion homeostasis
Trace metal homeostasis
Amino acids transporter
Bactoprenol synthesis
Methylglyoxal pathway
Respiration
Publications describing SubtiWiki and SubtiPathways
Raphael H Michna, Fabian M Commichau, Dominik Tödter, Christopher P Zschiedrich, Jörg Stülke
SubtiWiki-a database for the model organism Bacillus subtilis that links pathway, interaction and expression information.
Nucleic Acids Res: 2014, 42(Database issue);D692-8
[PubMed:24178028]
[WorldCat.org]
[DOI]
(I p)
Ulrike Mäder, Arne G Schmeisky, Lope A Flórez, Jörg Stülke
SubtiWiki--a comprehensive community resource for the model organism Bacillus subtilis.
Nucleic Acids Res: 2012, 40(Database issue);D1278-87
[PubMed:22096228]
[WorldCat.org]
[DOI]
(I p)
Christoph R Lammers, Lope A Flórez, Arne G Schmeisky, Sebastian F Roppel, Ulrike Mäder, Leendert Hamoen, Jörg Stülke
Connecting parts with processes: SubtiWiki and SubtiPathways integrate gene and pathway annotation for Bacillus subtilis.
Microbiology (Reading): 2010, 156(Pt 3);849-859
[PubMed:19959575]
[WorldCat.org]
[DOI]
(I p)