Difference between revisions of "NicK"
(→References) |
(→References) |
||
| Line 118: | Line 118: | ||
=References= | =References= | ||
| − | + | ==Reviews== | |
| + | <pubmed> 19943906 </pubmed> | ||
| + | ==Original Publications== | ||
<pubmed>17693500, 19943900, </pubmed> | <pubmed>17693500, 19943900, </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 11:04, 1 December 2009
- Description: DNA relaxase, similar to transposon protein
| Gene name | nicK |
| Synonyms | ydcR |
| Essential | no |
| Product | DNA relaxase, similar to transposon protein |
| Function | conjugation of ICE BS1 |
| MW, pI | 40 kDa, 5.131 |
| Gene length, protein length | 1056 bp, 352 aa |
| Immediate neighbours | ydcQ , ydcS |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 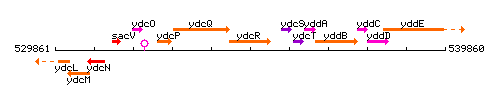
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU04870
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: plasmid replication initiation factor family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: P96635
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Elisabeth Grohmann
Autonomous plasmid-like replication of Bacillus ICEBs1: a general feature of integrative conjugative elements?
Mol Microbiol: 2010, 75(2);261-3
[PubMed:19943906]
[WorldCat.org]
[DOI]
(I p)
Original Publications