Difference between revisions of "PhrC"
(→References) |
(→Extended information on the protein) |
||
| Line 86: | Line 86: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' [[RapC]]-[[PhrC]] {{PubMed|12950917}} | + | * '''[[SubtInteract|Interactions]]:''' |
| + | ** [[RapC]]-[[PhrC]] {{PubMed|12950917}} | ||
| − | * '''Localization:''' | + | * '''[[Localization]]:''' |
=== Database entries === | === Database entries === | ||
Revision as of 10:22, 2 November 2011
- Description: response regulator aspartate phosphatase (RapC) regulator / competence and sporulation stimulating factor (CSF)
| Gene name | phrC |
| Synonyms | |
| Essential | no |
| Product | phosphatase (RapC) regulator / competence and sporulation stimulating factor (CSF) |
| Function | control of ComA activity |
| Interactions involving this protein in SubtInteract: PhrC | |
| MW, pI | 4 kDa, 8.16 |
| Gene length, protein length | 120 bp, 40 aa |
| Immediate neighbours | rapC, yczM |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 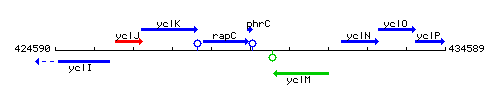
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
genetic competence, transcription factors and their control, quorum sensing, short peptides
This gene is a member of the following regulons
CcpA regulon, CodY regulon, ComA regulon, SigH regulon
The gene
Basic information
- Locus tag: BSU03780
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: binds to RapC, this results in the inability of RapC to interact with ComA PubMed
- Protein family: phr family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P94416
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Elizabeth Anne Shank, Roberto Kolter
Extracellular signaling and multicellularity in Bacillus subtilis.
Curr Opin Microbiol: 2011, 14(6);741-7
[PubMed:22024380]
[WorldCat.org]
[DOI]
(I p)
Original publications
Kassem Hamze, Daria Julkowska, Sabine Autret, Krzysztof Hinc, Krzysztofa Nagorska, Agnieszka Sekowska, I Barry Holland, Simone J Séror
Identification of genes required for different stages of dendritic swarming in Bacillus subtilis, with a novel role for phrC.
Microbiology (Reading): 2009, 155(Pt 2);398-412
[PubMed:19202088]
[WorldCat.org]
[DOI]
(P p)
Mridula Pottathil, April Jung, Beth A Lazazzera
CSF, a species-specific extracellular signaling peptide for communication among strains of Bacillus subtilis and Bacillus mojavensis.
J Bacteriol: 2008, 190(11);4095-9
[PubMed:18375560]
[WorldCat.org]
[DOI]
(I p)
Jennifer M Auchtung, Catherine A Lee, Alan D Grossman
Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides.
J Bacteriol: 2006, 188(14);5273-85
[PubMed:16816200]
[WorldCat.org]
[DOI]
(P p)
Natalia Comella, Alan D Grossman
Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis.
Mol Microbiol: 2005, 57(4);1159-74
[PubMed:16091051]
[WorldCat.org]
[DOI]
(P p)
Leighton Core, Marta Perego
TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis.
Mol Microbiol: 2003, 49(6);1509-22
[PubMed:12950917]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
A Yazgan, G Ozcengiz, M A Marahiel
Tn10 insertional mutations of Bacillus subtilis that block the biosynthesis of bacilysin.
Biochim Biophys Acta: 2001, 1518(1-2);87-94
[PubMed:11267663]
[WorldCat.org]
[DOI]
(P p)
B A Lazazzera, I G Kurtser, R S McQuade, A D Grossman
An autoregulatory circuit affecting peptide signaling in Bacillus subtilis.
J Bacteriol: 1999, 181(17);5193-200
[PubMed:10464187]
[WorldCat.org]
[DOI]
(P p)