Difference between revisions of "CspD"
| Line 118: | Line 118: | ||
** subject to Clp-dependent proteolysis upon glucose starvation {{PubMed|17981983}} | ** subject to Clp-dependent proteolysis upon glucose starvation {{PubMed|17981983}} | ||
** belongs to the 100 [[most abundant proteins]] {{PubMed|15378759}} | ** belongs to the 100 [[most abundant proteins]] {{PubMed|15378759}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 5135 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 13748 {{PubMed|24696501}} | ||
=Biological materials = | =Biological materials = | ||
Revision as of 09:40, 17 April 2014
- Description: cold shock protein
| Gene name | cspD |
| Synonyms | |
| Essential | no |
| Product | cold shock protein |
| Function | RNA chaperone |
| Gene expression levels in SubtiExpress: cspD | |
| MW, pI | 7 kDa, 4.31 |
| Gene length, protein length | 198 bp, 66 aa |
| Immediate neighbours | ugtP, degR |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed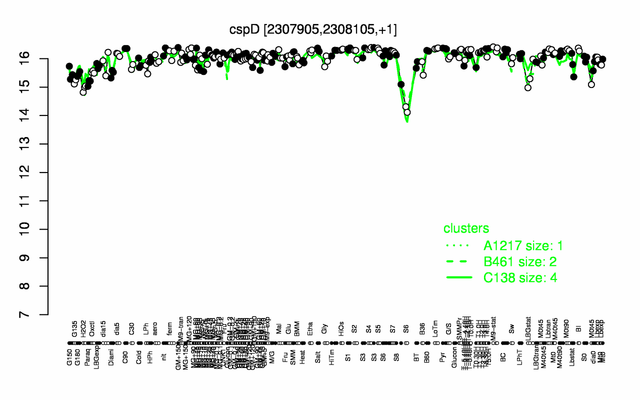
| |
Contents
Categories containing this gene/protein
RNA chaperones, cold stress proteins, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU21930
Phenotypes of a mutant
Database entries
- BsubCyc: BSU21930
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- BsubCyc: BSU21930
- Structure:
- UniProt: P51777
- KEGG entry: [2]
- E.C. number:
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Operon:
- Regulation:
- weakly induced by cold shock PubMed
- Regulatory mechanism:
- Additional information:
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
- belongs to the 100 most abundant proteins PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 5135 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 13748 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications
Ulf Gerth, Holger Kock, Ilja Kusters, Stephan Michalik, Robert L Switzer, Michael Hecker
Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis.
J Bacteriol: 2008, 190(1);321-31
[PubMed:17981983]
[WorldCat.org]
[DOI]
(I p)
Christine Eymann, Annette Dreisbach, Dirk Albrecht, Jörg Bernhardt, Dörte Becher, Sandy Gentner, Le Thi Tam, Knut Büttner, Gerrit Buurman, Christian Scharf, Simone Venz, Uwe Völker, Michael Hecker
A comprehensive proteome map of growing Bacillus subtilis cells.
Proteomics: 2004, 4(10);2849-76
[PubMed:15378759]
[WorldCat.org]
[DOI]
(P p)
Tanja Kaan, Georg Homuth, Ulrike Mäder, Julia Bandow, Thomas Schweder
Genome-wide transcriptional profiling of the Bacillus subtilis cold-shock response.
Microbiology (Reading): 2002, 148(Pt 11);3441-3455
[PubMed:12427936]
[WorldCat.org]
[DOI]
(P p)
Carsten L Beckering, Leif Steil, Michael H W Weber, Uwe Völker, Mohamed A Marahiel
Genomewide transcriptional analysis of the cold shock response in Bacillus subtilis.
J Bacteriol: 2002, 184(22);6395-402
[PubMed:12399512]
[WorldCat.org]
[DOI]
(P p)
Michael H W Weber, Ingo Fricke, Niclas Doll, Mohamed A Marahiel
CSDBase: an interactive database for cold shock domain-containing proteins and the bacterial cold shock response.
Nucleic Acids Res: 2002, 30(1);375-8
[PubMed:11752341]
[WorldCat.org]
[DOI]
(I p)
M H Weber, C L Beckering, M A Marahiel
Complementation of cold shock proteins by translation initiation factor IF1 in vivo.
J Bacteriol: 2001, 183(24);7381-6
[PubMed:11717297]
[WorldCat.org]
[DOI]
(P p)
M H Weber, A V Volkov, I Fricke, M A Marahiel, P L Graumann
Localization of cold shock proteins to cytosolic spaces surrounding nucleoids in Bacillus subtilis depends on active transcription.
J Bacteriol: 2001, 183(21);6435-43
[PubMed:11591689]
[WorldCat.org]
[DOI]
(P p)
T Schindler, P L Graumann, D Perl, S Ma, F X Schmid, M A Marahiel
The family of cold shock proteins of Bacillus subtilis. Stability and dynamics in vitro and in vivo.
J Biol Chem: 1999, 274(6);3407-13
[PubMed:9920884]
[WorldCat.org]
[DOI]
(P p)
P Graumann, T M Wendrich, M H Weber, K Schröder, M A Marahiel
A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures.
Mol Microbiol: 1997, 25(4);741-56
[PubMed:9379903]
[WorldCat.org]
[DOI]
(P p)