Difference between revisions of "CodY"
(→Labs working on this gene/protein) |
(→Original Publications) |
||
| Line 169: | Line 169: | ||
<pubmed>18083814,12618455, 23569278 24843172 </pubmed> | <pubmed>18083814,12618455, 23569278 24843172 </pubmed> | ||
==Original Publications== | ==Original Publications== | ||
| − | <pubmed>19542274,17493123,18641142,19202088,16995897,17993518,9287005,11331605,17218307,19500589,12591885,19202088,11331605,15228537,8793880, 15937175,15916605,15916606,11331605,15228537 19749041 7783641 20935095 21097623 21699902 21764931 22981860 22512862,21856856 23911932 24296669 24163341</pubmed> | + | <pubmed>19542274,17493123,18641142,19202088,16995897,17993518,9287005,11331605,17218307,19500589,12591885,19202088,11331605,15228537,8793880, 15937175,15916605,15916606,11331605,15228537 19749041 7783641 20935095 21097623 21699902 21764931 22981860 22512862,21856856 23911932 24296669 24163341 25157083 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 12:25, 19 September 2014
- Description: regulation of a large regulon (more than 100 genes and operons) in response to branched-chain amino acid limitation
| Gene name | codY |
| Synonyms | |
| Essential | no |
| Product | transcriptional pleiotropic repressor |
| Function | regulation of a large regulon in response to
branched-chain amino acid limitation to the presence of branched-chain amino acids |
| Gene expression levels in SubtiExpress: codY | |
| Interactions involving this protein in SubtInteract: CodY | |
| Metabolic function and regulation of this protein in SubtiPathways: codY | |
| MW, pI | 28 kDa, 4.75 |
| Gene length, protein length | 777 bp, 259 aa |
| Immediate neighbours | clpY, flgB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 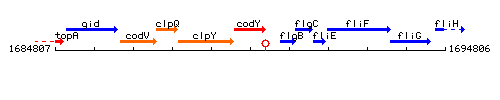
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed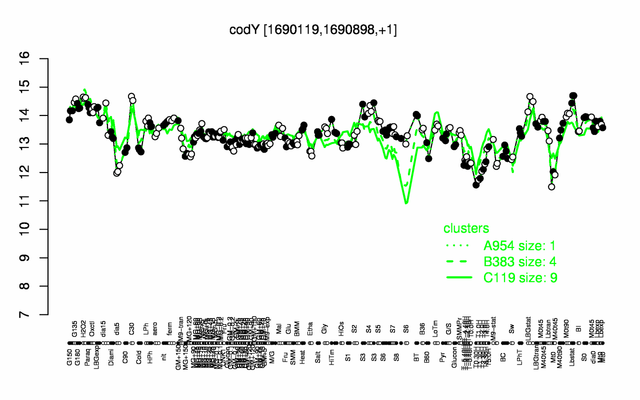
| |
Contents
Categories containing this gene/protein
transcription factors and their control, regulators of core metabolism, phosphoproteins
This gene is a member of the following regulons
The CodY regulon
The gene
Basic information
- Locus tag: BSU16170
Phenotypes of a mutant
- no swarming motility on B medium. PubMed
- the mutation suppresses the mucoid phenotype of motA or motB mutants due to loss of DegU phosphorylation and concomitant reduced expression of the capB-capC-capA-capE operon PubMed
- inactivation of codY suppresses the requirement of a relA sasA sasB triple mutant for branched chain amino acids, methionine and threonine PubMed
Database entries
- BsubCyc: BSU16170
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: codY family (according to Swiss-Prot)
- Paralogous protein(s):
Genes/ operons controlled by CodY
Extended information on the protein
- Kinetic information:
- Domains: contains a GAF domain (ligand binding domain)
- Modification: phosphorylation on Ser-215 PubMed
- Effectors of protein activity: GTP and branched chained amino acids (BCAA) increase the affinity of CodY for its DNA target sequences PubMed
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU16170
- UniProt: P39779
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
- the intracellular concentration of CodY is about 2.5 myM (according to PubMed)
- number of protein molecules per cell (minimal medium with glucose and ammonium): 955 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 3409 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 350 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 351 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 272 PubMed
Biological materials
- Mutant:
- GP566, available in Jörg Stülke's lab
- a codY::erm mutant is available in Linc Sonenshein's lab
- a codY::spc (BB1043) mutant is available in Linc Sonenshein's, Fabian Commichau's and Jörg Stülke's labs
- Expression vector:
- for expression, purification in E. coli with N-terminal His-tag, in pWH844: pGP245, available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Linc Sonenshein, Tufts University, Boston, MA, USA Homepage
- Tony Wilkinson, York University, U.K. Homepage
- Oscar Kuipers, University of Groningen, The Netherlands, Homepage
Your additional remarks
References
Reviews
The CodY regulon
Original Publications
Yasutaro Fujita, Takenori Satomura, Shigeo Tojo, Kazutake Hirooka
CcpA-mediated catabolite activation of the Bacillus subtilis ilv-leu operon and its negation by either CodY- or TnrA-mediated negative regulation.
J Bacteriol: 2014, 196(21);3793-806
[PubMed:25157083]
[WorldCat.org]
[DOI]
(I p)
Jia Mun Chan, Sarah B Guttenplan, Daniel B Kearns
Defects in the flagellar motor increase synthesis of poly-γ-glutamate in Bacillus subtilis.
J Bacteriol: 2014, 196(4);740-53
[PubMed:24296669]
[WorldCat.org]
[DOI]
(I p)
Allison Kriel, Shaun R Brinsmade, Jessica L Tse, Ashley K Tehranchi, Alycia N Bittner, Abraham L Sonenshein, Jue D Wang
GTP dysregulation in Bacillus subtilis cells lacking (p)ppGpp results in phenotypic amino acid auxotrophy and failure to adapt to nutrient downshift and regulate biosynthesis genes.
J Bacteriol: 2014, 196(1);189-201
[PubMed:24163341]
[WorldCat.org]
[DOI]
(I p)
Baoping Ling, Min Sun, Siwei Bi, Zhihong Jing, Zhiguo Wang
Molecular dynamics simulations of isoleucine-release pathway in GAF domain of N-CodY from Bacillus Subtilis.
J Mol Graph Model: 2013, 44;232-40
[PubMed:23911932]
[WorldCat.org]
[DOI]
(I p)
Allison Kriel, Alycia N Bittner, Sok Ho Kim, Kuanqing Liu, Ashley K Tehranchi, Winnie Y Zou, Samantha Rendon, Rui Chen, Benjamin P Tu, Jue D Wang
Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance.
Mol Cell: 2012, 48(2);231-41
[PubMed:22981860]
[WorldCat.org]
[DOI]
(I p)
Andrea Wünsche, Elke Hammer, Maike Bartholomae, Uwe Völker, Andreas Burkovski, Gerald Seidel, Wolfgang Hillen
CcpA forms complexes with CodY and RpoA in Bacillus subtilis.
FEBS J: 2012, 279(12);2201-14
[PubMed:22512862]
[WorldCat.org]
[DOI]
(I p)
Shaun R Brinsmade, Abraham L Sonenshein
Dissecting complex metabolic integration provides direct genetic evidence for CodY activation by guanine nucleotides.
J Bacteriol: 2011, 193(20);5637-48
[PubMed:21856856]
[WorldCat.org]
[DOI]
(I p)
Lewis V Wray, Susan H Fisher
Bacillus subtilis CodY operators contain overlapping CodY binding sites.
J Bacteriol: 2011, 193(18);4841-8
[PubMed:21764931]
[WorldCat.org]
[DOI]
(I p)
Boris R Belitsky, Abraham L Sonenshein
Roadblock repression of transcription by Bacillus subtilis CodY.
J Mol Biol: 2011, 411(4);729-43
[PubMed:21699902]
[WorldCat.org]
[DOI]
(I p)
Boris R Belitsky, Abraham L Sonenshein
Contributions of multiple binding sites and effector-independent binding to CodY-mediated regulation in Bacillus subtilis.
J Bacteriol: 2011, 193(2);473-84
[PubMed:21097623]
[WorldCat.org]
[DOI]
(I p)
Shaun R Brinsmade, Roelco J Kleijn, Uwe Sauer, Abraham L Sonenshein
Regulation of CodY activity through modulation of intracellular branched-chain amino acid pools.
J Bacteriol: 2010, 192(24);6357-68
[PubMed:20935095]
[WorldCat.org]
[DOI]
(I p)
Anuradha C Villapakkam, Luke D Handke, Boris R Belitsky, Vladimir M Levdikov, Anthony J Wilkinson, Abraham L Sonenshein
Genetic and biochemical analysis of the interaction of Bacillus subtilis CodY with branched-chain amino acids.
J Bacteriol: 2009, 191(22);6865-76
[PubMed:19749041]
[WorldCat.org]
[DOI]
(I p)
Heike Preis, Rita A Eckart, Rajani K Gudipati, Nadja Heidrich, Sabine Brantl
CodY activates transcription of a small RNA in Bacillus subtilis.
J Bacteriol: 2009, 191(17);5446-57
[PubMed:19542274]
[WorldCat.org]
[DOI]
(I p)
Vladimir M Levdikov, Elena Blagova, Vicki L Colledge, Andrey A Lebedev, David C Williamson, Abraham L Sonenshein, Anthony J Wilkinson
Structural rearrangement accompanying ligand binding in the GAF domain of CodY from Bacillus subtilis.
J Mol Biol: 2009, 390(5);1007-18
[PubMed:19500589]
[WorldCat.org]
[DOI]
(I p)
Kassem Hamze, Daria Julkowska, Sabine Autret, Krzysztof Hinc, Krzysztofa Nagorska, Agnieszka Sekowska, I Barry Holland, Simone J Séror
Identification of genes required for different stages of dendritic swarming in Bacillus subtilis, with a novel role for phrC.
Microbiology (Reading): 2009, 155(Pt 2);398-412
[PubMed:19202088]
[WorldCat.org]
[DOI]
(P p)
Shigeo Tojo, Takenori Satomura, Kanako Kumamoto, Kazutake Hirooka, Yasutaro Fujita
Molecular mechanisms underlying the positive stringent response of the Bacillus subtilis ilv-leu operon, involved in the biosynthesis of branched-chain amino acids.
J Bacteriol: 2008, 190(18);6134-47
[PubMed:18641142]
[WorldCat.org]
[DOI]
(I p)
Luke D Handke, Robert P Shivers, Abraham L Sonenshein
Interaction of Bacillus subtilis CodY with GTP.
J Bacteriol: 2008, 190(3);798-806
[PubMed:17993518]
[WorldCat.org]
[DOI]
(I p)
Wiep Klaas Smits, Tran Thu Hoa, Leendert W Hamoen, Oscar P Kuipers, David Dubnau
Antirepression as a second mechanism of transcriptional activation by a minor groove binding protein.
Mol Microbiol: 2007, 64(2);368-81
[PubMed:17493123]
[WorldCat.org]
[DOI]
(P p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Robert P Shivers, Sean S Dineen, Abraham L Sonenshein
Positive regulation of Bacillus subtilis ackA by CodY and CcpA: establishing a potential hierarchy in carbon flow.
Mol Microbiol: 2006, 62(3);811-22
[PubMed:16995897]
[WorldCat.org]
[DOI]
(P p)
Pascale Joseph, Manoja Ratnayake-Lecamwasam, Abraham L Sonenshein
A region of Bacillus subtilis CodY protein required for interaction with DNA.
J Bacteriol: 2005, 187(12);4127-39
[PubMed:15937175]
[WorldCat.org]
[DOI]
(P p)
Shigeo Tojo, Takenori Satomura, Kaori Morisaki, Josef Deutscher, Kazutake Hirooka, Yasutaro Fujita
Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA.
Mol Microbiol: 2005, 56(6);1560-73
[PubMed:15916606]
[WorldCat.org]
[DOI]
(P p)
Robert P Shivers, Abraham L Sonenshein
Bacillus subtilis ilvB operon: an intersection of global regulons.
Mol Microbiol: 2005, 56(6);1549-59
[PubMed:15916605]
[WorldCat.org]
[DOI]
(P p)
Robert P Shivers, Abraham L Sonenshein
Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids.
Mol Microbiol: 2004, 53(2);599-611
[PubMed:15228537]
[WorldCat.org]
[DOI]
(P p)
Hyun-Jin Kim, Sam-In Kim, Manoja Ratnayake-Lecamwasam, Kiyoshi Tachikawa, Abraham L Sonenshein, Mark Strauch
Complex regulation of the Bacillus subtilis aconitase gene.
J Bacteriol: 2003, 185(5);1672-80
[PubMed:12591885]
[WorldCat.org]
[DOI]
(P p)
M Ratnayake-Lecamwasam, P Serror, K W Wong, A L Sonenshein
Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels.
Genes Dev: 2001, 15(9);1093-103
[PubMed:11331605]
[WorldCat.org]
[DOI]
(P p)
L V Wray, A E Ferson, S H Fisher
Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H.
J Bacteriol: 1997, 179(17);5494-501
[PubMed:9287005]
[WorldCat.org]
[DOI]
(P p)
P Serror, A L Sonenshein
Interaction of CodY, a novel Bacillus subtilis DNA-binding protein, with the dpp promoter region.
Mol Microbiol: 1996, 20(4);843-52
[PubMed:8793880]
[WorldCat.org]
[DOI]
(P p)
F J Slack, P Serror, E Joyce, A L Sonenshein
A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon.
Mol Microbiol: 1995, 15(4);689-702
[PubMed:7783641]
[WorldCat.org]
[DOI]
(P p)