Difference between revisions of "YmdB"
(→Database entries) |
|||
| Line 60: | Line 60: | ||
* '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG13421] | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG13421] | ||
| + | |||
| + | * '''GEO entry:''' [http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39142 GSE39142] (''[[ymdB]] '' mutant vs. wild type) | ||
=== Additional information=== | === Additional information=== | ||
Revision as of 17:35, 14 December 2013
- Description: phosphodiesterase, controls bistable gene expression
| Gene name | ymdB |
| Synonyms | |
| Essential | no |
| Product | phosphodiesterase |
| Function | control of bistable gene expression |
| Gene expression levels in SubtiExpress: ymdB | |
| MW, pI | 29,1 kDa, 6.50 |
| Gene length, protein length | 792 bp, 264 amino acids |
| Immediate neighbours | rny, spoVS |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed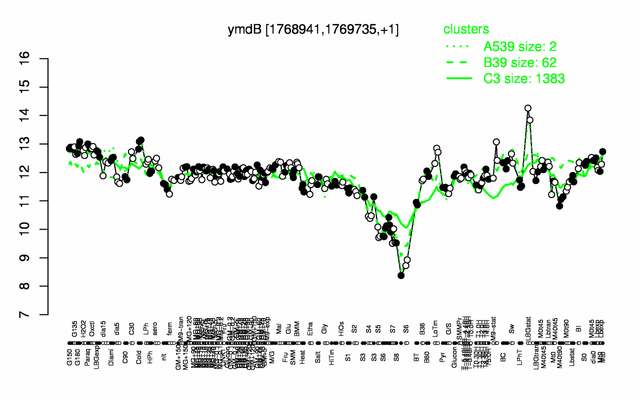
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU16970
Phenotypes of a mutant
- strong overexpression of hag PubMed
- defective in biofilm formation PubMed
- the phenotypes of the ymdB mutant can be suppressed by overexpression of slrR PubMed
- inactivation of ymdB restores beta-lactam resistance in a sigM mutant PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- phosphodiesterase activity toward 2',3'-cAMP PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- UniProt: O31775
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation: constitutive
- Regulatory mechanism:
- Additional information: there is a terminator between rny and ymdB, most transcripts terminate there PubMed
Biological materials
- Mutant:
- GP583 (spc), available in Jörg Stülke's lab PubMed
- GP922 (cat), available in Jörg Stülke's lab PubMed
- GP921 (spc) NCIB3610 derivate, available in Jörg Stülke's lab PubMed
- GP969 (ymdB(E39Q)-cat) inactive enzyme, available in Jörg Stülke's lab PubMed
- Expression vector:
- for expression/ purification from B. subtilis with N-terminal Strep-tag, for SPINE, in pGP380: pGP1041, available in Jörg Stülke's lab
- for expression/ purification from B. subtilis with C-terminal Strep-tag, for SPINE, in pGP382: pGP1919, available in Jörg Stülke's lab
- for expression/ purification from E. coli with N-terminal His-tag, in pWH844: pGP1040, available in Jörg Stülke's lab
- for expression/ purification from E. coli with N-terminal Strep-tag, in pGP172: pGP1917, available in Jörg Stülke's lab PubMed
- GP970 (ymdB-Strep (cat)), purification from B. subtilis, for SPINE, available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- FLAG-tag construct: GP1018 (spc, based on pGP1331), available in Jörg Stülke's lab
- Antibody:
Labs working on this gene/protein
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
Functional and structural analysis of orthologs in other organisms