Difference between revisions of "YvyD"
(→References) |
|||
| Line 58: | Line 58: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU35310&redirect=T BSU35310] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/yvyD.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/yvyD.html] | ||
| Line 95: | Line 96: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU35310&redirect=T BSU35310] | ||
* '''Structure:''' | * '''Structure:''' | ||
Revision as of 14:55, 2 April 2014
- Description: general stress protein, required for ribosome dimerization in the stationary phase, required for protection against paraquat stress
| Gene name | yvyD |
| Synonyms | yviI |
| Essential | no |
| Product | unknown |
| Function | dimerization of ribosomes in the stationary phase, protection against paraquat stress |
| Gene expression levels in SubtiExpress: yvyD | |
| MW, pI | 21 kDa, 5.184 |
| Gene length, protein length | 567 bp, 189 aa |
| Immediate neighbours | secA, smiA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 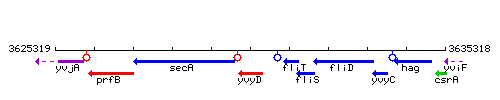
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed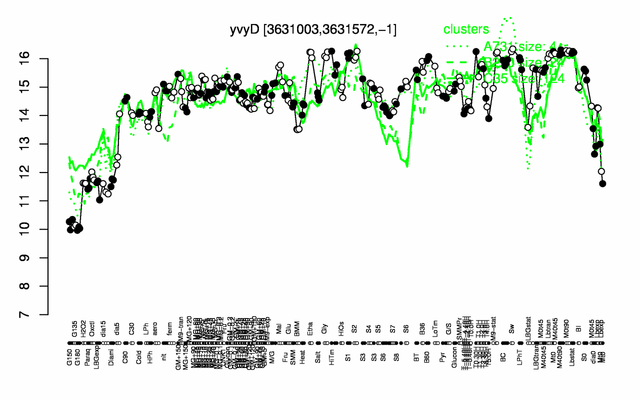
| |
Contents
Categories containing this gene/protein
general stress proteins (controlled by SigB), resistance against oxidative and electrophile stress, membrane proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU35310
Phenotypes of a mutant
- the mutant is cold-sensitive
- delayed recovery from stationary phase and delayed germination
Database entries
- BsubCyc: BSU35310
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ribosomal protein S30Ae family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- phosphorylated on Arg-6 PubMed
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
- membrane protein
Database entries
- BsubCyc: BSU35310
- Structure:
- UniProt: P28368
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: yvyD PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Publications on the homologous proteins from other bacteria
Pranav Puri, Thomas H Eckhardt, Linda E Franken, Fabrizia Fusetti, Marc C A Stuart, Egbert J Boekema, Oscar P Kuipers, Jan Kok, Bert Poolman
Lactococcus lactis YfiA is necessary and sufficient for ribosome dimerization.
Mol Microbiol: 2014, 91(2);394-407
[PubMed:24279750]
[WorldCat.org]
[DOI]
(I p)
Yury S Polikanov, Gregor M Blaha, Thomas A Steitz
How hibernation factors RMF, HPF, and YfiA turn off protein synthesis.
Science: 2012, 336(6083);915-8
[PubMed:22605777]
[WorldCat.org]
[DOI]
(I p)