SepF
- Description: part of the divisome, recruits FtsZ to the membrane
| Gene name | sepF |
| Synonyms | ylmF |
| Essential | no |
| Product | FtsZ-interacting protein |
| Function | recruitment of FtsZ |
| Gene expression levels in SubtiExpress: sepF | |
| Interactions involving this protein in SubtInteract: SepF | |
| MW, pI | 17 kDa, 4.863 |
| Gene length, protein length | 447 bp, 149 aa |
| Immediate neighbours | ylmE, ylmG |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 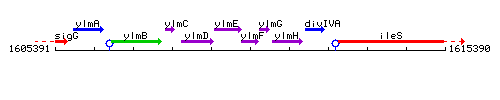
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
cell division, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU15390
Phenotypes of a mutant
- perturbation of the formation of properly formed division septa
- less efficient cell division results in longer cells. Electron microscopy reveals strongly distorted division septa.
- the sepF mutation in combination with a constitutively active form of WalR (WalR-R204C) results in the formation of cell wall-less L-forms PubMed
- the sepF mutation is synthetically lethal in combination with an ezrA mutation or an ftsA mutation
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: sepF family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: O31728
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Leendert Hamoen, CBCB, Newcastle University, UK
Shu Ishikawa, Nara Institute of Science and Technology, Nara, Japan
Your additional remarks
SepF mutation is synthetic lethal in combination with an ezrA mutation or an ftsA mutation.
References
Reviews
Kevin M Devine
Bacterial L-forms on tap: an improved methodology to generate Bacillus subtilis L-forms heralds a new era of research.
Mol Microbiol: 2012, 83(1);10-3
[PubMed:22126136]
[WorldCat.org]
[DOI]
(I p)
David W Adams, Jeff Errington
Bacterial cell division: assembly, maintenance and disassembly of the Z ring.
Nat Rev Microbiol: 2009, 7(9);642-53
[PubMed:19680248]
[WorldCat.org]
[DOI]
(I p)
Original Publications