Difference between revisions of "RsbX"
| Line 1: | Line 1: | ||
| − | + | * '''Description:''' [[Protein kinases and phosphatases|protein serine phosphatase]], feedback PP2C, dephosphorylates [[RsbS]] and [[RsbR]]<br/><br/> | |
| − | |||
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
| Line 39: | Line 38: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | + | <br/><br/> | |
| − | |||
| − | |||
| − | |||
| − | |||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
| Line 121: | Line 116: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=rsbX_523650_524249_1 rsbX] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=rsbX_523650_524249_1 rsbX] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' |
** ''[[rsbR]]'': [[SigA]] [http://www.ncbi.nlm.nih.gov/sites/entrez/8002610 PubMed] | ** ''[[rsbR]]'': [[SigA]] [http://www.ncbi.nlm.nih.gov/sites/entrez/8002610 PubMed] | ||
** ''[[rsbV]]:'' [[SigB]] [http://www.ncbi.nlm.nih.gov/pubmed/11544224 PubMed] | ** ''[[rsbV]]:'' [[SigB]] [http://www.ncbi.nlm.nih.gov/pubmed/11544224 PubMed] | ||
| Line 152: | Line 147: | ||
=References= | =References= | ||
| − | '''Additional publications:''' {{PubMed|21362065}} | + | '''Additional publications:''' {{PubMed|23320651,21362065}} |
<pubmed>8002610,8682769,9658013,8002609,10671474,9068644, 19923733,11544224, 15466036 8824586 </pubmed> | <pubmed>8002610,8682769,9658013,8002609,10671474,9068644, 19923733,11544224, 15466036 8824586 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 12:31, 5 February 2013
- Description: protein serine phosphatase, feedback PP2C, dephosphorylates RsbS and RsbR
| Gene name | rsbX |
| Synonyms | |
| Essential | no |
| Product | protein serine phosphatase, feedback PP2C |
| Function | control of SigB activity |
| Gene expression levels in SubtiExpress: rsbX | |
| Interactions involving this protein in SubtInteract: RsbX | |
| Metabolic function and regulation of this protein in SubtiPathways: Stress | |
| MW, pI | 21 kDa, 6.23 |
| Gene length, protein length | 597 bp, 199 aa |
| Immediate neighbours | sigB, ydcF |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 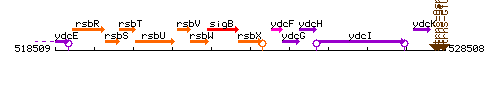
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
protein modification, sigma factors and their control, general stress proteins (controlled by SigB)
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU04740
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P17906
- KEGG entry: [3]
- E.C. number: 3.1.3.3
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Bill Haldenwang, San Antonio, USA
- Chet Price, Davis, USA homepage
Your additional remarks
References
Additional publications: PubMed
Masatoshi Suganuma, Aik Hong Teh, Masatomo Makino, Nobutaka Shimizu, Tomonori Kaneko, Kunio Hirata, Masaki Yamamoto, Takashi Kumasaka
Crystallization and preliminary X-ray analysis of the stress-response PPM phosphatase RsbX from Bacillus subtilis.
Acta Crystallogr Sect F Struct Biol Cryst Commun: 2009, 65(Pt 11);1128-30
[PubMed:19923733]
[WorldCat.org]
[DOI]
(I p)
Chien-Cheng Chen, Michael D Yudkin, Olivier Delumeau
Phosphorylation and RsbX-dependent dephosphorylation of RsbR in the RsbR-RsbS complex of Bacillus subtilis.
J Bacteriol: 2004, 186(20);6830-6
[PubMed:15466036]
[WorldCat.org]
[DOI]
(P p)
A Petersohn, M Brigulla, S Haas, J D Hoheisel, U Völker, M Hecker
Global analysis of the general stress response of Bacillus subtilis.
J Bacteriol: 2001, 183(19);5617-31
[PubMed:11544224]
[WorldCat.org]
[DOI]
(P p)
J M Scott, T Mitchell, W G Haldenwang
Stress triggers a process that limits activation of the Bacillus subtilis stress transcription factor sigma(B).
J Bacteriol: 2000, 182(5);1452-6
[PubMed:10671474]
[WorldCat.org]
[DOI]
(P p)
N Smirnova, J Scott, U Voelker, W G Haldenwang
Isolation and characterization of Bacillus subtilis sigB operon mutations that suppress the loss of the negative regulator RsbX.
J Bacteriol: 1998, 180(14);3671-80
[PubMed:9658013]
[WorldCat.org]
[DOI]
(P p)
U Voelker, T Luo, N Smirnova, W Haldenwang
Stress activation of Bacillus subtilis sigma B can occur in the absence of the sigma B negative regulator RsbX.
J Bacteriol: 1997, 179(6);1980-4
[PubMed:9068644]
[WorldCat.org]
[DOI]
(P p)
X Yang, C M Kang, M S Brody, C W Price
Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor.
Genes Dev: 1996, 10(18);2265-75
[PubMed:8824586]
[WorldCat.org]
[DOI]
(P p)
A Dufour, U Voelker, A Voelker, W G Haldenwang
Relative levels and fractionation properties of Bacillus subtilis σ(B) and its regulators during balanced growth and stress.
J Bacteriol: 1996, 178(13);3701-9 sigma
[PubMed:8682769]
[WorldCat.org]
[DOI]
(P p)
A A Wise, C W Price
Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor sigma B in response to environmental signals.
J Bacteriol: 1995, 177(1);123-33
[PubMed:8002610]
[WorldCat.org]
[DOI]
(P p)
U Voelker, A Dufour, W G Haldenwang
The Bacillus subtilis rsbU gene product is necessary for RsbX-dependent regulation of sigma B.
J Bacteriol: 1995, 177(1);114-22
[PubMed:8002609]
[WorldCat.org]
[DOI]
(P p)